In a latest research printed within the journal Nature, a big group of researchers from america (U.S.) used single-cell ribonucleic acid (RNA) sequencing mixed with high-resolution fluorescence in situ hybridization to find out the identities of the assorted cell sorts that coordinate spatially to offer rise to the complicated morphological construction of the center.
 Examine: Spatially organized mobile communities kind the growing human coronary heart. Picture Credit score: beerkoff / Shutterstock
Examine: Spatially organized mobile communities kind the growing human coronary heart. Picture Credit score: beerkoff / Shutterstock
Background
Every complicated construction of the human coronary heart has particular roles that contribute to environment friendly cardiac perform, and the interruption of any of those features can result in congenital disabilities equivalent to congenital coronary heart illness in youngsters and cardiac illnesses equivalent to valvulopathies and cardiomyopathies in adults. Nevertheless, regardless of the important position of the center within the human physique, the group and performance of the cardiac buildings and the way they work together with each other stay poorly understood.
Concerning the research
Within the current research, the researchers used a single-cell RNA sequencing (scRNAseq) strategy together with multiplexed error-robust fluorescence in situ hybridization (MER-FISH). This technique allowed them to mix single-cell transcriptomes and spatial biology and visualize, analyze, and quantify the RNA transcripts of numerous genes from a single cell.
They started by figuring out the cell lineages that have been a part of the growing coronary heart, which helped decide how the assorted cardiac cell sorts assemble into complicated buildings and coordinate to control the perform of the human coronary heart. The scRNAseq was performed in replicates and analyzed for human hearts in numerous phases of development, ranging from 9 weeks and going as much as 16 weeks post-conception.
The obtained single cells, over 140 million in quantity, have been transcriptionally categorized into 5 cell compartments: cardiomyocytes, endothelial, mesenchymal, neuronal, and blood. Inside these cell compartments, evaluation of gene markers helped establish 12 cell lessons, with subsequent clustering analyses figuring out 39 populations and 75 subpopulations of cells.
MER-FISH was then used to spatially map the center cells and discover the mobile mechanisms by which the reworking and morphogenesis of the center, together with the ventricular wall growth, have been directed. The group of the cells recognized utilizing scRNAseq, particularly throughout developmental durations such because the compaction of the myocardial wall, was explored utilizing MER-FISH imaging.
The research then aimed to decipher the meeting of those particular cardiovascular cells into the mobile neighborhoods that come collectively to kind the multi-cell buildings that contribute to coronary heart perform. The scientists additionally explored the organizational and mobile complexity of particular areas, such because the ventricles, by exploring the cells throughout the ventricles that have been recognized, remoted, and mapped utilizing MER-FISH. Moreover, mouse fashions have been used to interrogate the interactions between cells by in vivo experiments, and pluripotent stem cells from people have been used to guage the identical in in vitro experiments.
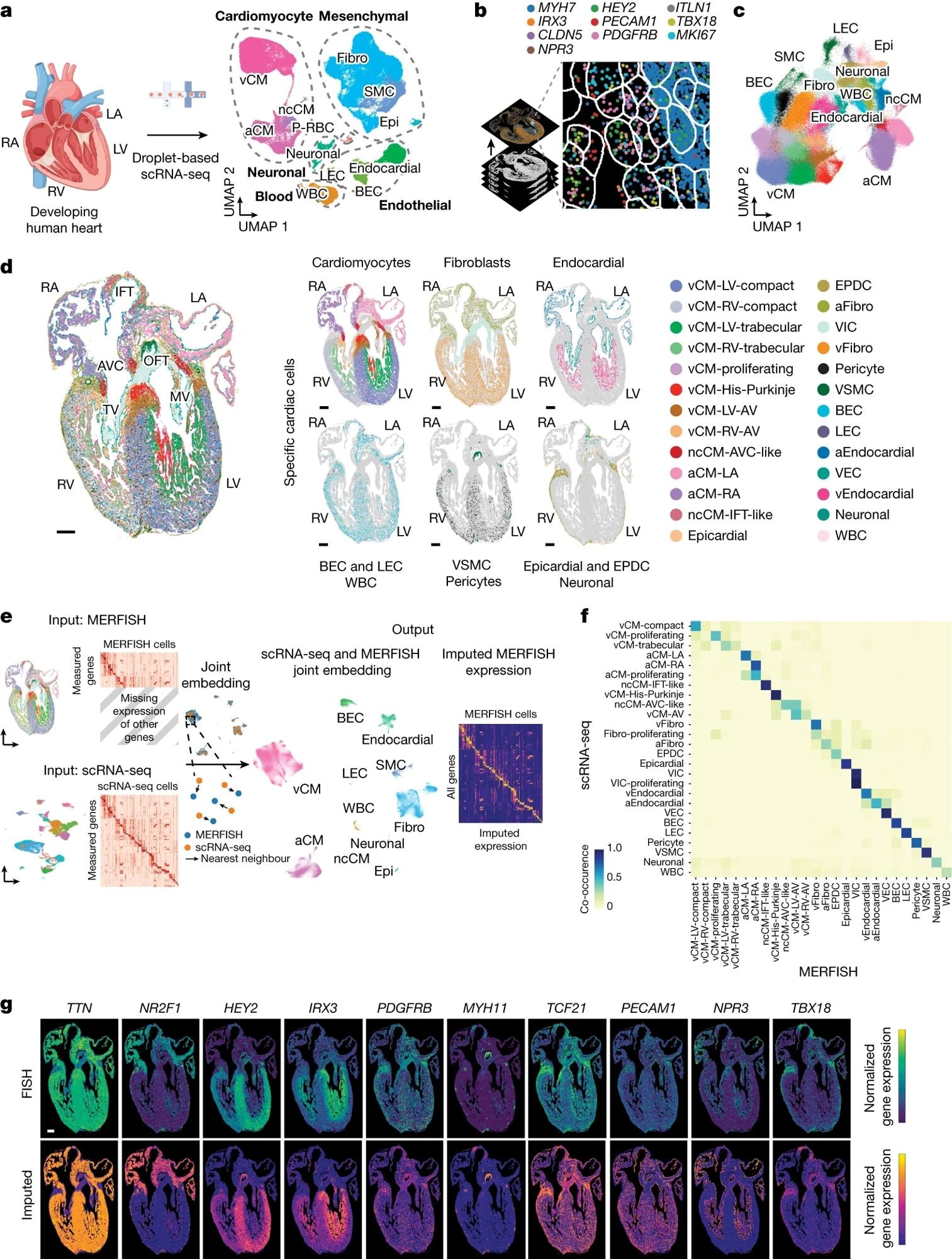 a, Left, schematic of experiment. Proper, scRNA-seq identifies a various vary of distinct cardiac cells that create the growing human coronary heart as displayed by uniform manifold approximation and projection (UMAP) of ~143,000 cells. b, Schematic exhibits how 238 cardiac-cell-specific genes have been spatially recognized utilizing MERFISH. Pseudo-coloured dots mark the situation of particular person molecules of ten particular RNA transcripts. c, Roughly 250,000 MERFISH-identified cardiac cells have been clustered into particular cell populations as proven by UMAP and colored accordingly in d. d, Recognized MERFISH cells have been spatially mapped throughout a frontal part of a 13 p.c.w. coronary heart (left) and proven in accordance with main cell lessons (proper). e, Joint embedding between MERFISH and age-matched scRNA-seq datasets enabled cell label switch and MERFISH gene imputation. f, Co-occurrence heatmap exhibits the correspondence of cell annotations of MERFISH cells to these transferred from the 13 p.c.w. scRNA-seq dataset. g, Gene imputation efficiency was validated spatially by evaluating normalized gene expression profiles of marker genes measured by MERFISH with the corresponding imputed gene expression profiles. Epi, epicardial; MV, mitral valve; P–RBC, platelet–crimson blood cell; TV, tricuspid valve. Scale bar, 250 µm (g). Illustration in a was created utilizing BioRender (https://www.biorender.com).
a, Left, schematic of experiment. Proper, scRNA-seq identifies a various vary of distinct cardiac cells that create the growing human coronary heart as displayed by uniform manifold approximation and projection (UMAP) of ~143,000 cells. b, Schematic exhibits how 238 cardiac-cell-specific genes have been spatially recognized utilizing MERFISH. Pseudo-coloured dots mark the situation of particular person molecules of ten particular RNA transcripts. c, Roughly 250,000 MERFISH-identified cardiac cells have been clustered into particular cell populations as proven by UMAP and colored accordingly in d. d, Recognized MERFISH cells have been spatially mapped throughout a frontal part of a 13 p.c.w. coronary heart (left) and proven in accordance with main cell lessons (proper). e, Joint embedding between MERFISH and age-matched scRNA-seq datasets enabled cell label switch and MERFISH gene imputation. f, Co-occurrence heatmap exhibits the correspondence of cell annotations of MERFISH cells to these transferred from the 13 p.c.w. scRNA-seq dataset. g, Gene imputation efficiency was validated spatially by evaluating normalized gene expression profiles of marker genes measured by MERFISH with the corresponding imputed gene expression profiles. Epi, epicardial; MV, mitral valve; P–RBC, platelet–crimson blood cell; TV, tricuspid valve. Scale bar, 250 µm (g). Illustration in a was created utilizing BioRender (https://www.biorender.com).
Outcomes
The findings revealed that numerous cardiac cell sorts belonged to particular subpopulations that have been a part of particular communities, with the practical specialization outlined in accordance with the anatomical area by which they have been current and the mobile ecosystem. The cardiomyocyte lineages have been the biggest cell compartment recognized utilizing MER-FISH. The research additionally discovered that cells that belonged to non-cardiomyocyte cell compartments additionally underwent segregation into populations and subpopulations and contributed to the formation of particular buildings and areas of the center.
Cardiomyocyte subpopulations within the ventricular areas exhibited a capability to assemble complicated laminal buildings within the ventricular wall and kind mobile communities with different subpopulations of cardiac cells. Moreover, the in vivo and in vitro experiments performed to know the interactions between cells revealed that the spatial group of subpopulations of cardiac cells in the course of the morphogenesis of the ventricular wall was performed by numerous signaling pathways.
The research additionally discovered cardiac areas composed of spatially organized combos of cell populations that segregated collectively, referred to as mobile communities. These mobile communities assorted within the quantity and forms of cell populations, and inside these communities, neighbors of every cardiac cell inside a 150-micrometer radius have been outlined. These interacting populations of cells additionally had distinct mobile signaling pathways.
Conclusions
General, the research discovered that cardiomyocytes have been the biggest compartment of cell sorts within the growing coronary heart, and all of the cell sorts exhibited distinct structural and regional distributions within the coronary heart. Particular cell populations additionally shaped mobile communities in numerous combos, with signaling pathways between the cell populations throughout the group defining their construction and performance. The research helped in understanding the event of the complicated construction of the human coronary heart, offering potential avenues to deal with structural coronary heart illnesses.
Journal reference:
- Farah, E.N., Hu, R.Okay., Kern, C., Zhang, Q., Lu, T., Ma, Q., Tran, S., Zhang, B., Carlin, D., Monell, A., Blair, A.P., Wang, Z., Eschbach, J., Li, B., Destici, E., Ren, B., Evans, S.M., Chen, S., Zhu, Q. and Chi, N.C. (2024). Spatially organized mobile communities kind the growing human coronary heart. Nature. DOI: 10.1038/s4158602407171z, https://www.nature.com/articles/s41586-024-07171-z
Supply hyperlink